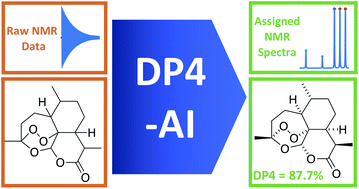
Every organic chemist has had to solve problems of structure elucidation, such as determining the structure of a biologically-active natural product or understanding the products of a reaction. These problems are often difficult and may be a bottleneck of chemical discovery. Structural misassignment leads to the waste of time and resources. In the last two decades computational tools have become increasingly useful in tackling these problems, with the DP4 Probability developed by the Goodman Lab being a key contribution to this toolkit (https://doi.org/10.1021/ja105035r). By comparing experimental NMR spectra and those computed for candidate structures, DP4 quantifies confidence in structural assignment, enabling chemists to use their resources more effectively.
Kris: Five years ago when I joined Jonathan’s group calculating a DP4 probability involved lots of spreadsheets and the time-consuming task of managing computer time effectively. I decided that the major task of automating all the complex parts of the process would be difficult, but would lead to a process which was much less laborious and immune to operational errors. This turned out to be much more difficult than I anticipated. Each step of the process that was automated made testing easier and faster. This meant I could discover new problems more quickly. Fortunately, a Master’s student then joined the project.

Alex: I had the right combination of bravery and naïvety to agree to take on the project. By the end of my Master’s project, we had a proof-of-concept program for carbon NMR. The structure elucidation performance wasn’t great, but all the moving bits were there: raw NMR data and DFT computational data went in, and a structure assignment came out. I then decided to take on a PhD project in the same area, and continued the process of discovering new challenges in NMR automation.

Kris

Alex
We faced several key problems in the transition from proof-of-concept to a robust and general process. The first was obtaining the raw NMR FID data for testing, as it is not available from scientific literature. Several synthetic groups generously provided the data, often requiring searching through dusty lab books and long forgotten flash drives. Chemical research community need to think more carefully about how we can make this precious raw NMR data more discoverable and accessible.
We found that every component in the NMR processing workflow is crucial: if either the Fourier transform, peak picking or integration is imperfect, the errors cascade and the resulting structure assignment performance falls dramatically. We also had to be creative in getting the computer algorithms to think more like human chemists. Often, while staring at a spectrum the computer had very obviously assigned incorrectly, we would ask ourselves, "what was our thought process in determining that this assignment is wrong?" which we would encode as probabilities, matrices and scores.

Ultimately we were satisfied with the process and DP4-AI was born: structure elucidation from raw NMR data now works just as well as with manually interpreted NMR spectra. Human interaction has now been removed from DP4 calculations and the process is, much, much faster. Starting from just candidate structures and NMR data a DP4 probability calculation used to take about a day of human time, now it takes just one minute! DP4 calculations have been dramatically accelerated and the scope of potential applications expanded significantly.
We have tried as much as possible to ensure that DP4-AI is not only an interesting proof-of-concept, but a robust, general, accessible and user-friendly tool. The paper is open-access (https://doi.org/10.1039/D0SC00442A) and the software is open-source and available on GitHub (https://github.com/KristapsE/DP4-AI ). All the raw NMR and computational data used in the development and testing are also freely available (https://www.repository.cam.ac.uk/handle/1810/303161).
We hope DP4-AI and its components will be useful for integration into other automated NMR analysis workflows. We are keen to develop this idea further in various directions, but we are just as excited to see what other people will do with it!
Kris Ermanis and Alex Howarth





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in